Fetal, Pregnant Women and Infants Numerical Models

The models distributed in this project are licensed under a Creative Commons Attribution-Noncommercial-Share Alike 2.0 France License.
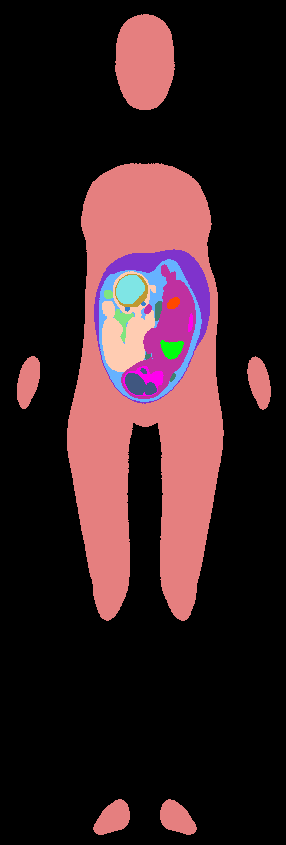
General Overview
The aim of both FEMONUM and then FETUS projects is to develop reliable 3D models to be used for numerical dosimetry studies. These models were built using different approaches combining image segmentation, growth modeling, smooth meshes deformation and physical deformation. The broad ideas of the approaches developped during the course of these two projects are summarized next.Ultrasound image segmentation
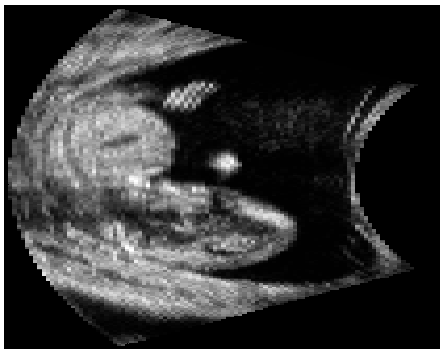
A database of 3D ultrasound image volumes acquired during the first trimester of pregnancy was provided by collaborating obstetricians, (Hospital Beaujon, Paris, FRANCE). Two main methods have been developed to cope with the need to differentiate fetal from maternal tissues on 3D ultrasound data to segment fetal body.- The first one aimed at separating amniotic fluid from fetal and maternal tissues.
An automated segmentation framework based on statistical homogeneity measures was designed to perform this task.
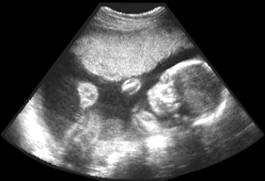
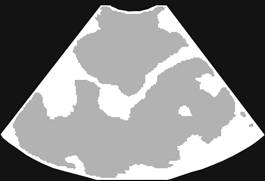
3D Ultrasound image
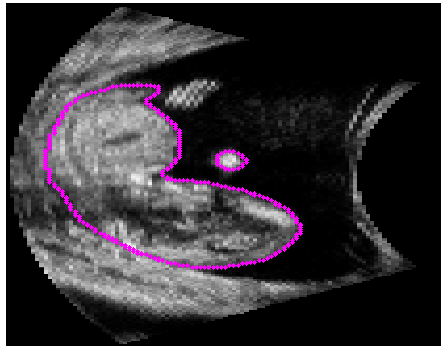
Segmentation
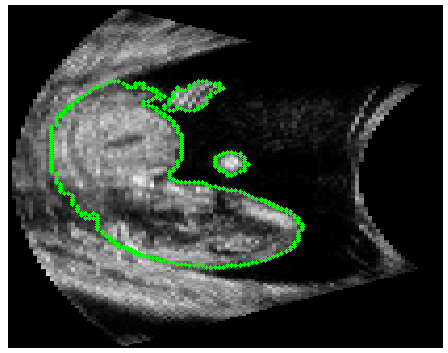
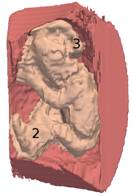
3D reconstruction of the fetus body
As a second step, segmentation errors and connections between the fetus and the uterine wall were manually corrected to generate a final mesh model.
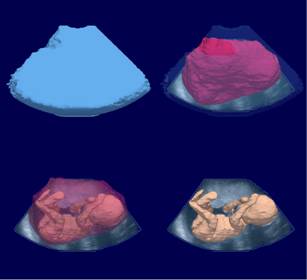
(blue) field of view, (pink) uterine internal wall,
(yellow) fetus body.
- The second method proposes a variational segmentation framework combining shape priors and parametric intensity distribution modeling for extracting the fetal envelope. Three types of information are injected in the segmentation process: tissue-specific parametric modeling of pixel intensities (determined automatically), a shape prior for the fetal envelope and a shape model of the fetus' back. The shape prior is encoded with Legendre moments and used to constraint the evolution of a level-set function. The back model is used to post-process the segmented fetal envelope.
|
|
|
|
|
3D Ultrasound image |
Manual Segmentation |
Automatic Segmentation |
MRI image segmentation
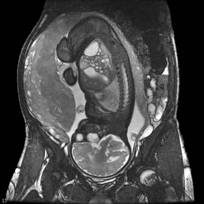
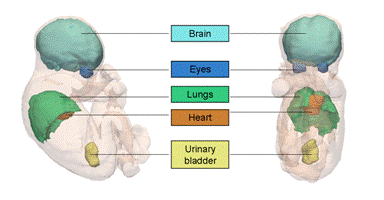
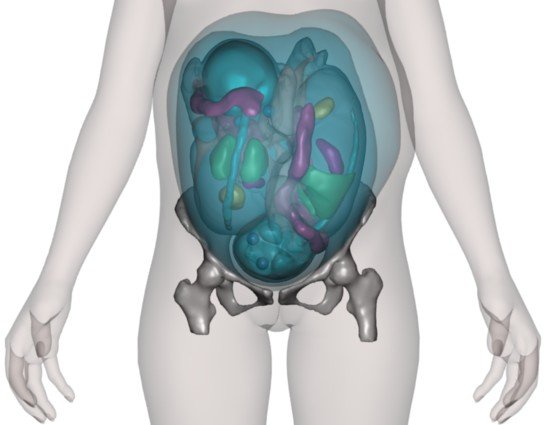
MRI image data provide a clear visualization of several maternal abdominal tissues (skin, bone, fat, muscles,...) and of fetal organs (body envelope, brain, lungs, heart,...).
In collaboration with the pediatric radiology department of the hospital Saint Vincent de Paul (Paris, France), we first performed a study to define the best suited MRI protocol for the segmentation and modeling of the fetus. The Steady State Free Precession was selected, offering the best image quality for the segmentation task.
A methodological framework was then developed for automated segmentation of several fetal structures, including the eyes, the brain-skull content, the spinal cord, the urinary bladder, and the fetal envelope. The segmentation process is based on the extraction of landmark points based on appearance models, iterative orientation of the fetus based on the landmark points and fine segmentation of tissue interfaces with a contrast-sensitive graph-cut partitioning of image data, within narrow bands of image data.
|
|
|
|
SSFP MRI image of a fetus |
3D fetus segmentation |
Growth modeling
A 3D articulated fetal growth model covering the main phases of pregnancy was developed. The structures of interest: skeleton, body envelope, brain and lungs were automatically or semi- automatically (depending on the stage of pregnancy) segmented from a database of images and surface meshes were generated. A generic mesh of each structure has been deformed towards the segmented ones. By interpolating linearly between fetal structures, each one can be generated at any age and in any position. This process results in an automated model, the operator being only required to specify tthe age and position of the desired estimated fetus.

Pregnant Woman Body modeling
Victoria is a non-gravid woman body envelope mesh model developed by Daz Studio, which can be manipulated and animated with the software Blender.
We used Victoria's body envelope to generate hybrid models of pregrant women:
- For models based on 3D ultrasound data, the abdominal surface of Victoria was not modified.
- For models based on MRI data, the abdominal surface of Victoria was deformed applying a series of automatically computed and controlled deformations using a physics-based interactive modeler.
|
|
|
||
|
Mesh-based pregnant woman model (fetal model based on MRI data). |
|||
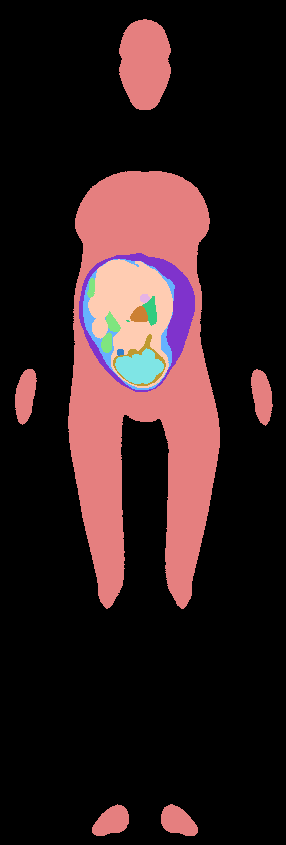
Pixel-based models were finally generated, at a desired spatial resolution, with different labels assigned to the different tissues.
|
|
|
||
|
Pixel-based pregnant woman model |
|||